Simplify discovery,
collaborate anywhere.
One tool to manage, explore, annotate, and collaborate on your large microscopy data.
Explore and Collaborate on your Microscopy Datasets from anywhere. Simplified discovery.
All the performance. None of the HURDLES.
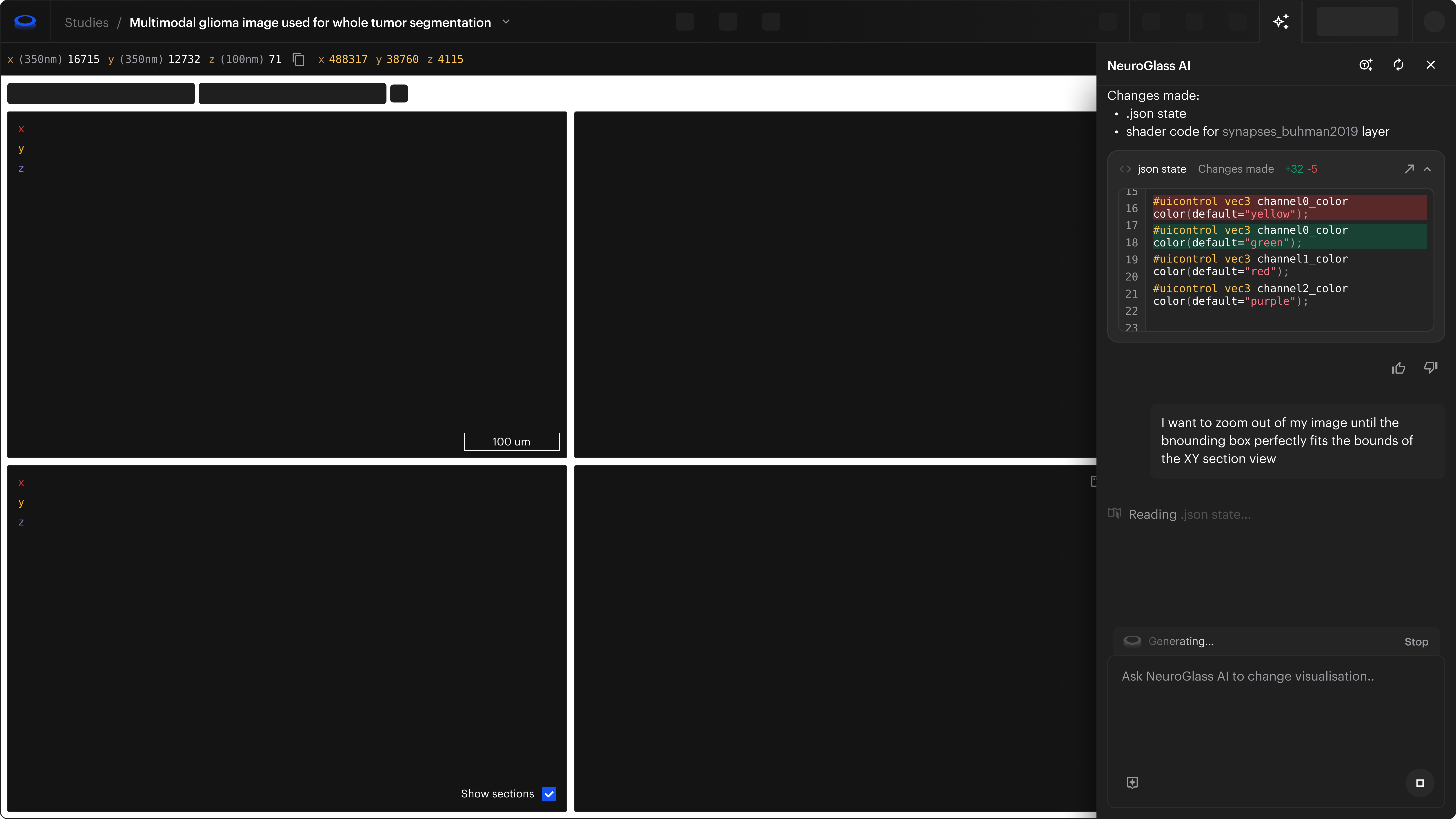
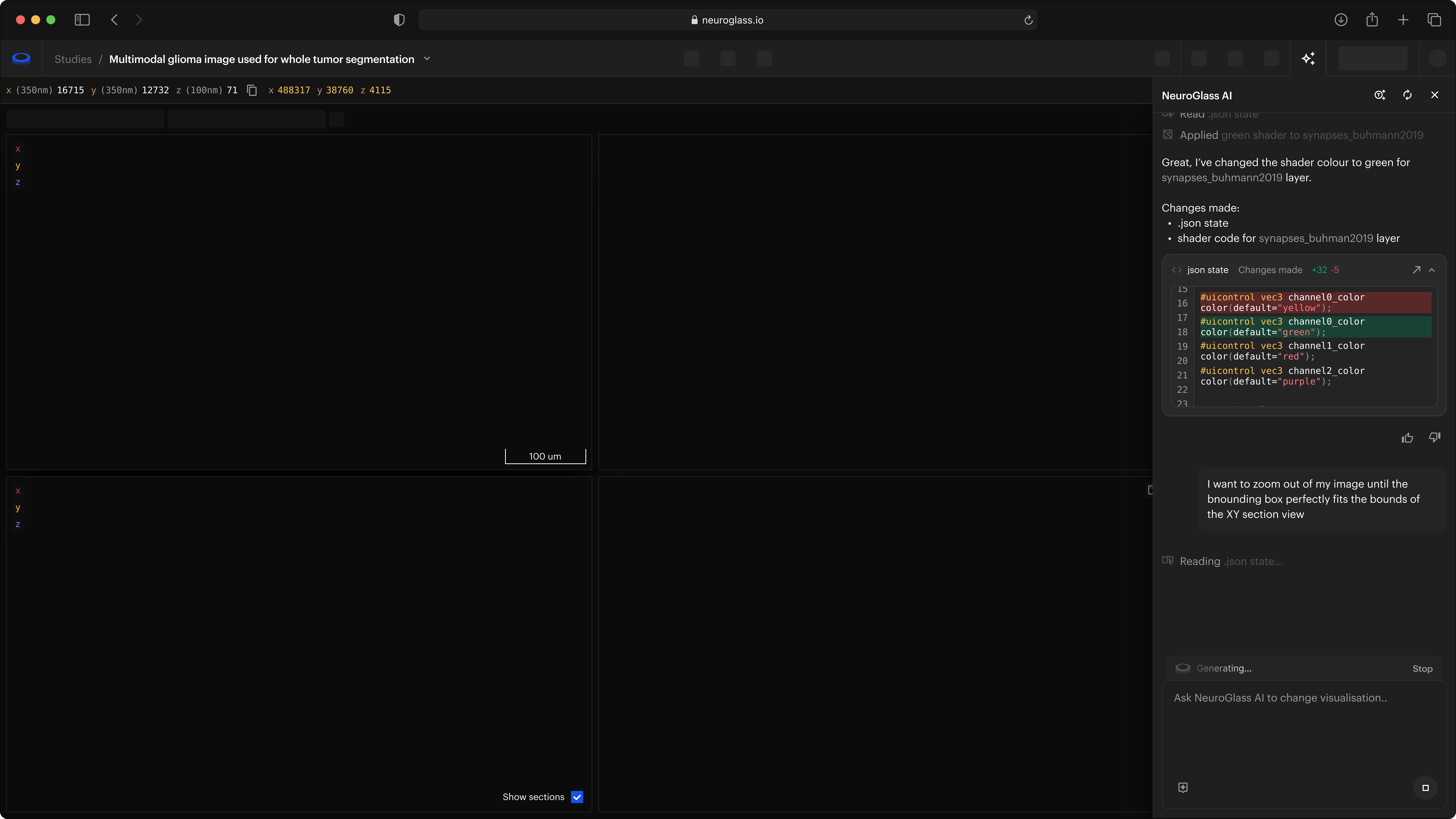
A novel Neuroglancer experience
Attributions: Zheng et al., Cell 2018; Buhmann et al., Nature Methods 2021; Heinrich et al., arXiv 2018
The fastest visualization engine for microscopy data
All the tools and performances of Neuroglancer with an improved user interface and experience.

.gif)
Powerful history
Keep track of everything you do, restore or go back to any previous state. It’s called reproducible science.

Figures. Capture key visualizations from your study
Highlight critical findings and share them with colleagues. Edit and refine whenever you want.

Figure 3 Segmentation of Adult Drosophila Melanogaster
Glances. Reproducible results
Capture a specific moment as you work on your study, preserving key insights for easy reference. Share a reproducible view of your data, enabling collaboration and research continuity.

Designed for your large scale volumetric 3D dataset

Connectomics

Cryo-EM/ET

Expansion Microscopy

High-Res Cellular Imaging

Developmental Imaging

Molecular Imaging
Electron Microscopy
Fluorescence Microscopy
Attributions
The MICrONS Consortium (Bae et al.) (2025), "Functional connectomics spanning multiple areas of mouse visual cortex.", Nature
Sedzicki, J., Ni, D., Lehmann, F. et al. Mechanism of cyclic β-glucan export by ABC transporter Cgt of Brucella. Nat Struct Mol Biol 29, 1170–1177 (2022). https://doi.org/10.1038/s41594-022-00868-7
Image set BBBC031v1 [Szkalisity et al. 2017], available from the Broad Bioimage Benchmark Collection [Ljosa et al., Nature Methods, 2012].
Richard Allen (University of Hawaii) (2011) CIL:27152, Paramecium multimicronucleatum, cell by organism, eukaryotic cell, Eukaryotic Protist, Ciliated Protist. CIL. Dataset. https://doi.org/doi:10.7295/W9CIL27152
Image set BBBC024vl [Svoboda David, Kozubkek Michal, Stejskal Stanislav. Generation of Digital Phantoms of Cell Nuclei and Simulation of Image Formation in 3D Image Cytometry. Cytometry Part A, John Wiley & Sons, Inc., 75A, 6, pp. 494-509, 16 pages. ISSN 1552-4922. 2009.] from the Broad Bioimage Benchmark Collection.
Walther N, Hossain MJ, Politi AZ, Koch B, Kueblbeck M, Ødegård-Fougner Ø, Lampe M, Ellenberg J. A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J Cell Biol. 2018 Jul 2;217(7):2309-2328. doi: 10.1083/jcb.201801048. Epub
Clarity does NOt stop at the image.
Keep track of your data and find what you need
Studies
Organize and visualize your experiments
Visualization engine
All the power of Neuroglancer with an improved user interface and usability experience.
Powerful history
Keep track of everything you do, go back to any previous state. It’s called reproducible science.
Glances
Crystallize any moment and share it seamlessly with other researchers.
Figures
Move, refine, snap. Store and organize the figures for your publications. And bring them back to life with a single click so you can tweak them and refine them.
Glances
Share and publish reproducible results
History
Decide whether NeuroGlass should automatically remember all your activities or if you prefer to manually save specific moments of your study.
Memory
NeuroGlass will remember all the settings and visualisation properties of your navigation in a Glance, so you can go back to it or share it with your collaborators
Share
When you share a Glance, your colleagues will see the exact view that you crafted for them
Publish
Does everyone agree that this Glance is worth sharing with the scientific community? Turn it into a Figure, download it, and include it in your next publication.

Figures
Capture key visualizations from your study
Create
Find the combination of visualisation settings that looks best for you, and create a high resolution snapshot ready for publication.
Edit and re-create
Do you need to make a small change? Every Figure is linked to a Glance, so you can go back to it, edit it and re-create a new version.
Bulk-edits
Coming soon.
Download
Once you are happy with your work, export your high resolution Figure.

Data Collections
Coming soon
Import and organize your datasets
Data schema flexibility
Import the metadata that your community is familiar with. You can add or remove fields to existing data schemas, or create your own!
Easy to use
NeuroGlass will generate a template for you to populate with your data, and re-upload.
Bulk-connect your experiments
All the experiments imported are connected together and managed for you. No more copy and paste.
Speed and scalability
Simplify bulk edits across studies, glances, and figures by applying visualisation settings to all related studies.
Attributions: Zheng et al., Cell 2018; Buhmann et al., Nature Methods 2021; Heinrich et al., arXiv 2018
Metadata
Link your metadata with native OME support or create custom metadata that fits your experiment.

Tagging
Group your Studies, Figures, and Glances with tags for quick, easy access whenever you need them.

Extensive search
Easily find your annotations, layers, Studies and more. NeuroGlass search through your whole database to find what you need.

Data Collections Ingestion & Validation
Import and organize your datasets. Select your experiment type, then link your dataset and metadata seamlessly with native OME support.

NeuroGlass for oRGANIZATIONs
Make it easy for your team to collaborate and publish
NeuroGlass works at scale, removing technical barriers from scientific Studies so your team can focus on the real challenges.

Easily share data within your institution, at scale
Remove the friction, let your team collaborate on annotations the easy way
Make your institution’s data available to the scientific community, as soon as you are ready
Customize NeuroGlass with the logo and colors of your organization
Gather usage statistics for your published Studies
Use NeuroGlass from our own secure deployment or on premise
Use your own fork of Neuroglancer, if needed
Work together. Discover more.
Collaboration should not be hard
Share everything you need with collaborators: simultaneously work on data, looks, annotations, and more
Short URLs
Your Studies can be shared easily with short and manageable links.
Easily share
You can share any content with your colleagues, whether they are NeuroGlass users or not.

Real-time concurrent edits
Collaborate on the same Study in real time—share ideas and get inspired.

Automate. Integrate. Innovate.
Built for scientists. Ready for developers.
Seamlessly import your data or programmatically create your Studies with our developer-friendly APIs.
Visit API documentation

Your data, protected by design.
Research securely. Collaborate confidently.
We take your data security seriously — that’s why we ensure it never leaves its trusted home.
Secure hosting
Whether deployed in your institution or on MetaCell Cloud, your research data never leaves your servers.
GDPR compliance
Your information is safe with us. Visit the Privacy Terms to know more about how we treat personal information.
Secure Single sign-on (SSO)
Integrate NeuroGlass with your authentication provider for easy access.

What is coming next
We are actively working to improve NeuroGlass. Want to fill in the blocks below?
Let us know what features you'd like to see
Collaborative annotations
OME metadata support
DOI support
Bulk updates to figures
Frequently Asked Questions
Can't find an answer? Contact us
What is NeuroGlass?
NeuroGlass is a cloud application that allows smooth visualisation of large microscopy data (e.g. Electron Microscopy, Light Sheet, etc). It also integrates an advanced UI that allows to organise and manage your data, to share it with your colleagues and to generate figures for your publication. All of this without the need to install anything on your computer.
What data does NeuroGlass support?
Neuroglass supports the OME-Zarr (NGFF), Zarr, n5, Single NIfTI image, Neuroglancer Precomputed, and Deep Zoom Image file formats. Neuroglass also supports data from the BOSS, DVID, and Render databases/APIs.
How do I import my data into NeuroGlass
You can import data into NeuroGlass in two ways: 1) Import a Neuroglancer link or 2) Create a Data Collection
When I create a Data Collection, do I need to upload the raw image files?
No, your data stays on your pre-existing cloud storage. During the Data Collection creation process, you will be asked to indicate the location of where your data is stored, and NeuroGlass will take care of the rest!
Is my data secure in NeuroGlass?
Yes, your data will not leave your pre-existing cloud storage. Metadata stored in NeuroGlass will be saved on MetaCell Cloud following best in class security standards. You’ll be able to delete it anytime, no question asked.
Can I collaborate with my team using NeuroGlass?
Yes, you can share your data with selected collaborators (regardless of whether they have an account on Neuroglass or not) or with anyone who has the link to your data.
Can I use NeuroGlass free of charge?
Yes, individual users can create an account and a few Studies and Figures for free. If you would like your team to take advantage of unlimited features, import data from your institution's bucket, and receive support check out our Pricing page or reach out to us.
Contributing back to the Neuroglancer community
We built NeuroGlass to accelerate research breakthroughs by simplifying collaboration and enhancing productivity. NeuroGlass wouldn't exist without Neuroglancer and the incredible contributions of its community.
NeuroGlass is committed to giving back—keeping all Neuroglancer improvements open source and freely available for everyone.
NeuroGlass is committed to giving back—keeping all Neuroglancer improvements open source and freely available for everyone.